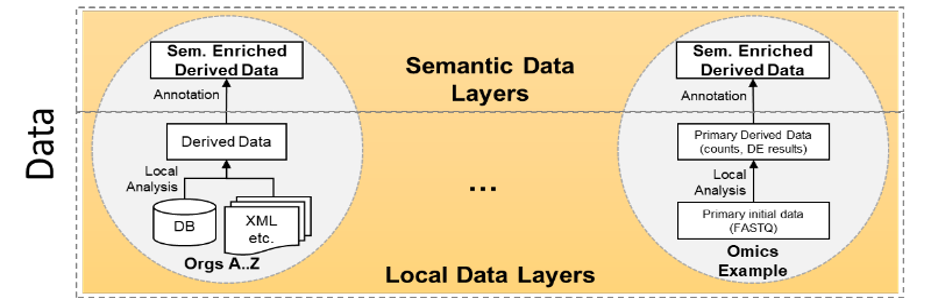
The main purpose of a semantic data layer is to provide meaningful semantics to data, to enable the connection between primary and secondary data. Another key aspect of the introduced semantic layer is that it stores derived data, which represents the analyzed and, in some cases, interpreted data by experts. Under "semantic data layer" we understand data structure for a single type of data such as RNA sequencing (RNA-seq), CAP gene expression analysis (CAGE) for RNA expression or whole exome sequencing (WES) for protein-coding genes (about 1% of the genome) in order to find mutation variants.
Semantic Data Layers
In the case of the RNA source data in FASTQ format, in a first step, counts of expressed RNA or small RNA are calculated. This is done with available tools such as OASISfor the calculation of small RNA counts. In a second step, differential expression scores (for e.g., p-values) between healthy and diseased subjects are calculated.
For data annotations the designed semantic layers utilize the controlled neuro-specific vocabularies and mappings available in the semantic lookup platform. The annotations are stored as key:value descriptions using controlled vocabulary for both key and value terms. Currently, the semantic lookup platform contains more than 20 pre-defined annotation keys, however, new (not normalized) keys are allowed as well.
| KEy | Ontology |
|---|---|
| Organism | NCBI Taxonomy |
| Tissue | BRENDA tissue/enzyme |
| Disease | Human Disease Ontology |
| Cell type | Cell Ontology |
| Cell line | Cell Line Ontology, Experimental Factor Ontology |
Example NGS: Terminologies used for semantic annotation of NGS Data