Prospective, longitudinal cohort studies, as have been started at DZNE, take years to enroll and follow up a substantial number of patients. In comparison, clinical routine data is the most comprehensive data source for phenotypic patient information. The memory clinic of the University Hospital of Bonn, for example, holds e-health records since 2000 with valuable longitudinal patient data that contain information about characteristics and progression of neurodegenerative diseases. Thus, if available and accessible, this real-life data can complement the cohort data collected at DZNE.
Incorporation of Longitudinal Electronic Patient Information
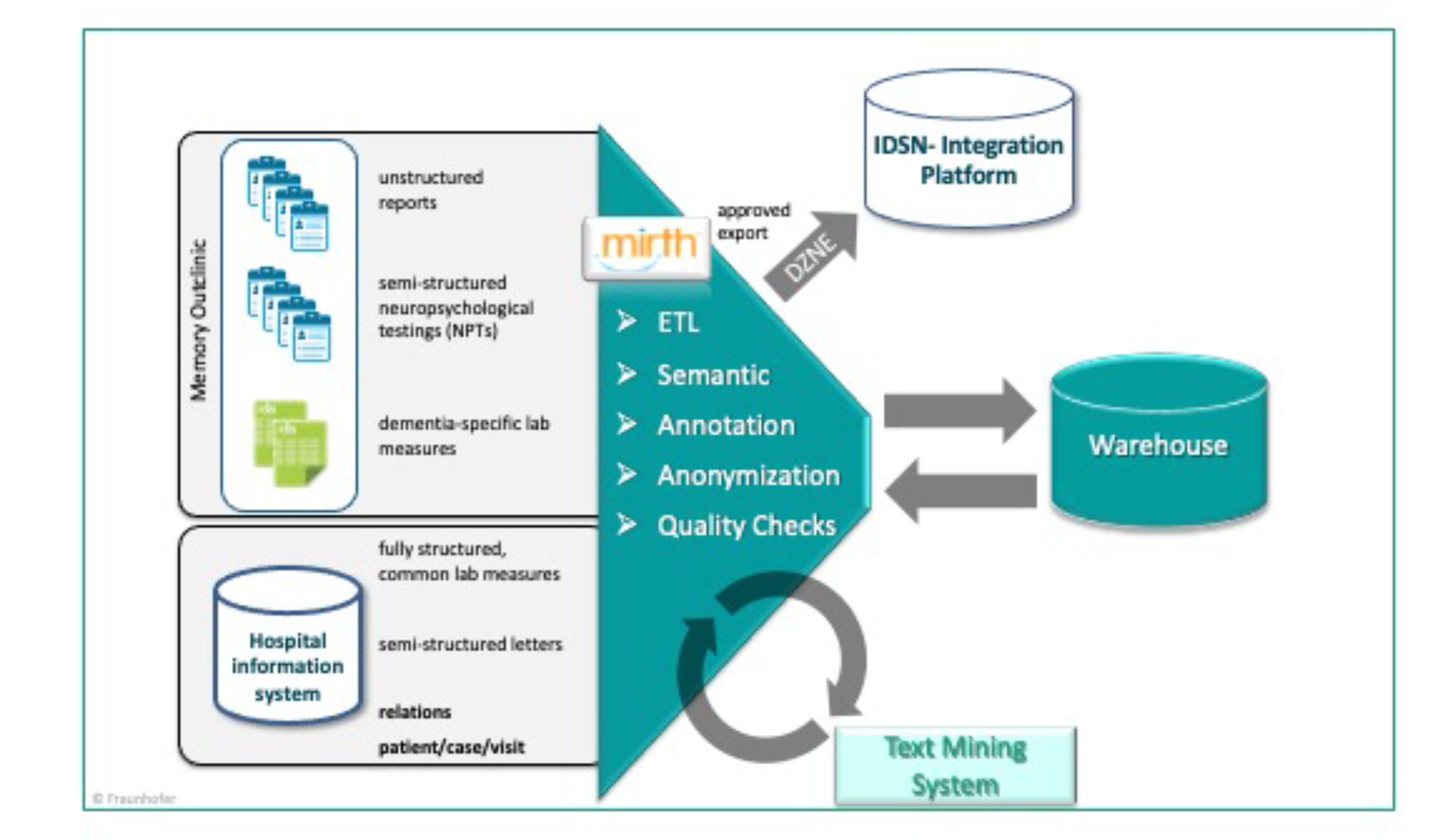
We set up an extract, load, and transform (ETL) process based on the software component Mirth Connect. All extracted information was semantically annotated and stored in a data warehouse. For further information extraction of unstructured and semi-structured resources a text mining system was integrated into the ETL process. The extracted and semantically annotated information was stored in the warehouse as well. After anonymization and data quality check an export file containing only structured information was generated. After approval through a data protection procedure the data was exported to IDSN clinical data base or integration platform.
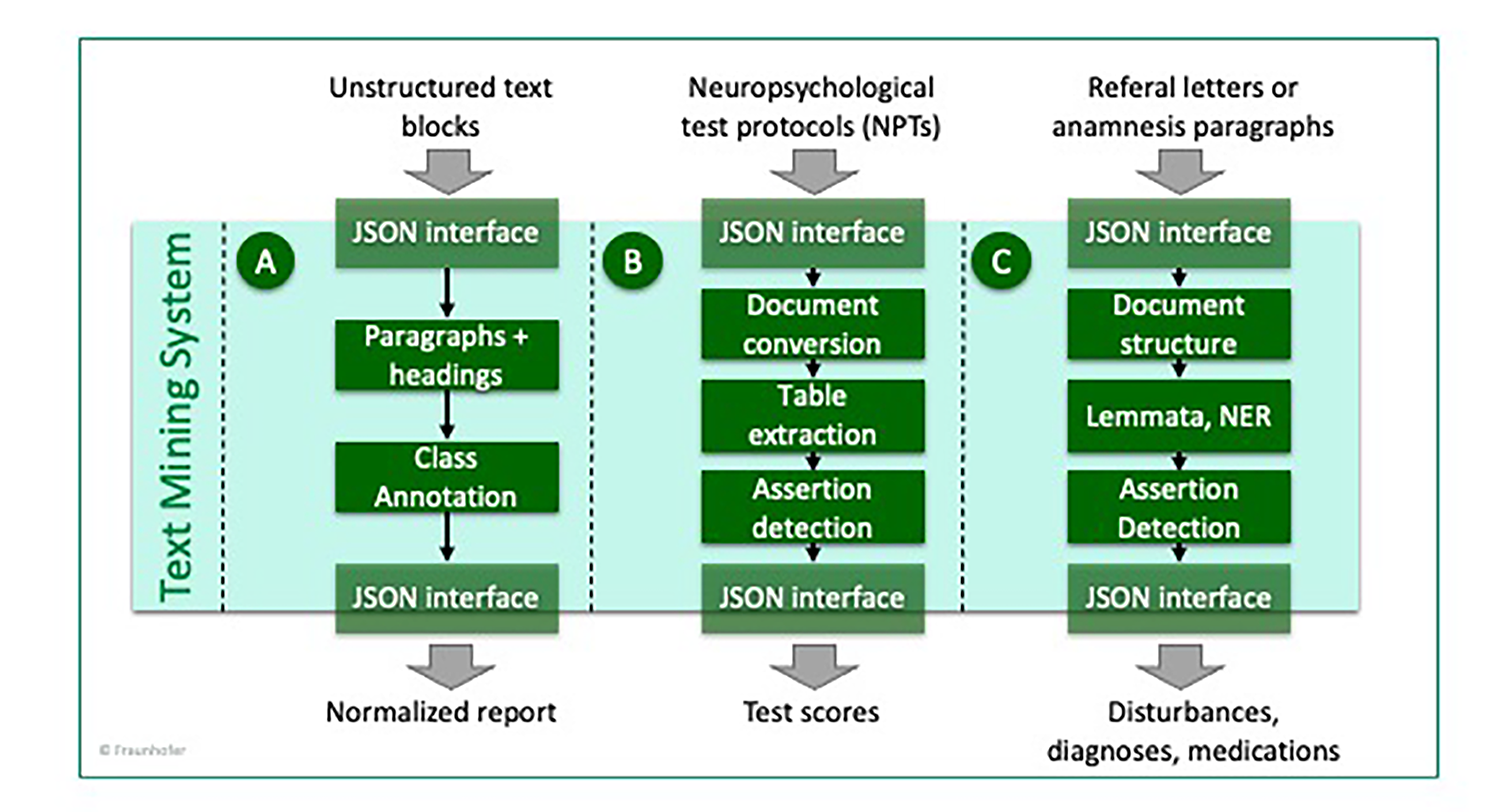
For information extraction from unstructured text blocks, such as neuropsychological test protocols, referral letters and anamnesis sections, we set up a text mining system with modular rule-based text mining workflows. Every module was developed while using a small, anonymized training data and evaluated on separate test sets. All modules can be easily re-used and adapted to new hospital settings.
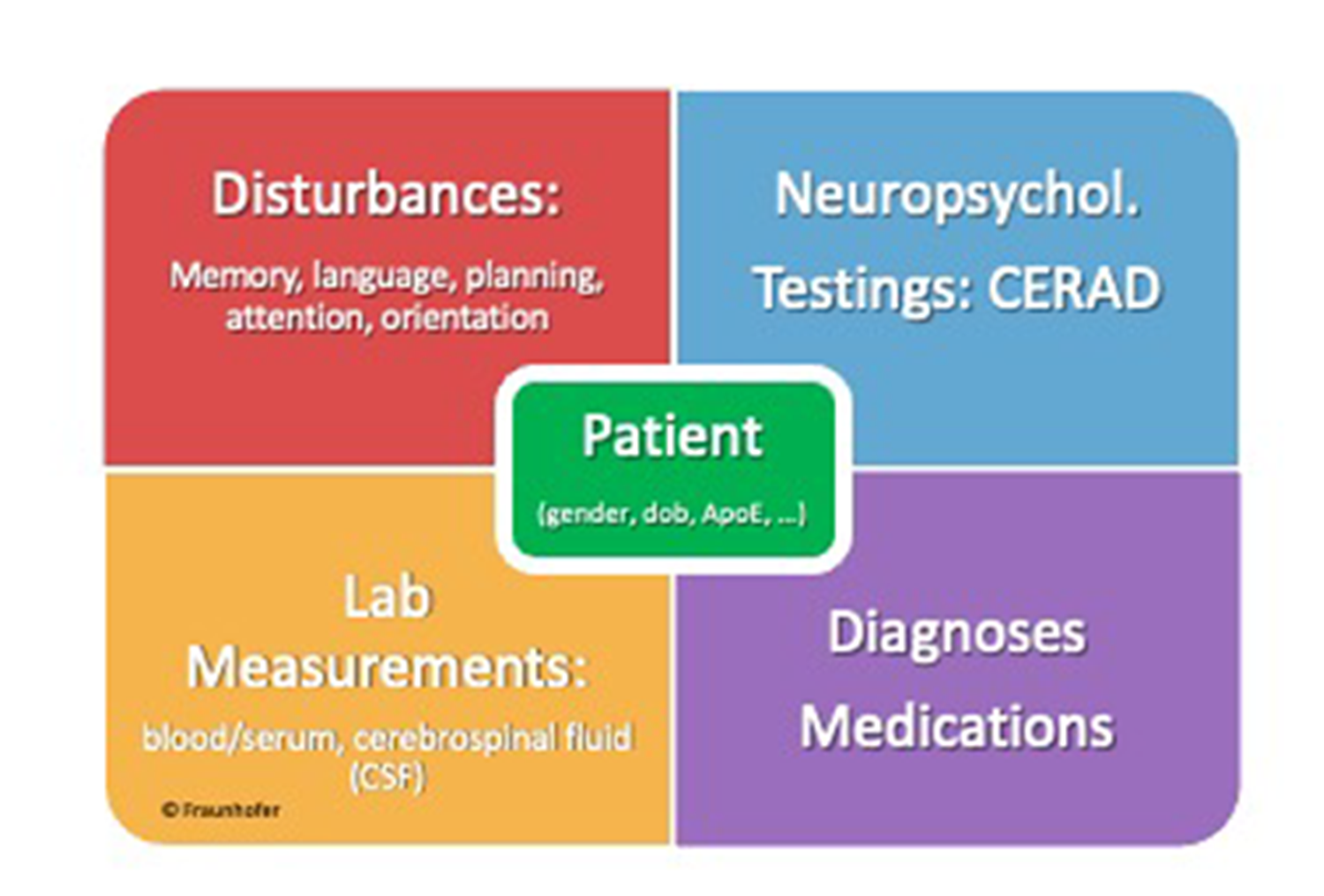
For semantic annotation of clinical neurodegeneration disorder data, we generated a clinical data model containing general patient master data, information about cognitive deficits, neuropsychological testing scores (such as CERAD), blood, serum and cerebrospinal fluid lab measurements, diagnosis, and medication
Overview of the overall information extraction process
We set up an extract, load, and transform (ETL) process based on the software component Mirth Connect. All extracted information was semantically annotated and stored in a data warehouse. For further information extraction of unstructured and semi-structured resources a text mining system was integrated into the ETL process. The extracted and semantically annotated information was stored in the warehouse as well. After anonymization and data quality check an export file containing only structured information was generated. After approval through a data protection procedure the data was exported to IDSN clinical data base or integration platform.
Overview of text mining system
For information extraction from unstructured text blocks, such as neuropsychological test protocols, referral letters and anamnesis sections, we set up a text mining system with modular rule-based text mining workflows. Every module was developed while using a small, anonymized training data and evaluated on separate test sets. All modules can be easily re-used and adapted to new hospital settings.
Semantic annotation of clinical data
For semantic annotation of clinical neurodegeneration disorder data, we generated a clinical data model containing general patient master data, information about cognitive deficits, neuropsychological testing scores (such as CERAD), blood, serum and cerebrospinal fluid lab measurements, diagnosis, and medication.